Chromatin remodeling
We are interested in the regulation of genome structure in eukaryotes by ATP-dependent chromatin remodeling complexes. These highly conserved “remodelers” are the only known factors that can directly alter the positioning of nucleosomes, the basic repeating unit of chromatin, comprising ~150 basepairs of DNA wrapped around a core of histone proteins. This activity is essential for cellular activities such as DNA replication, repair and transcription, which rely on access to functional DNA sequences that are rendered inaccessible within a nucleosome.
Remodelers contain a conserved ATPase domain that catalyzes the disruption of histone-DNA interactions, allowing for the histone core to be repositioned relative to the DNA. Association with different sets of domains and accessory factors allows remodelers to perform diverse functions, including sliding and ejecting the histone core, as well as exchanging histones in the nucleosome.
We are interested in understanding, at a mechanistic level, how the basic translocating activity of the remodeling ATPases is regulated by accessory domains and subunits to generate different products.
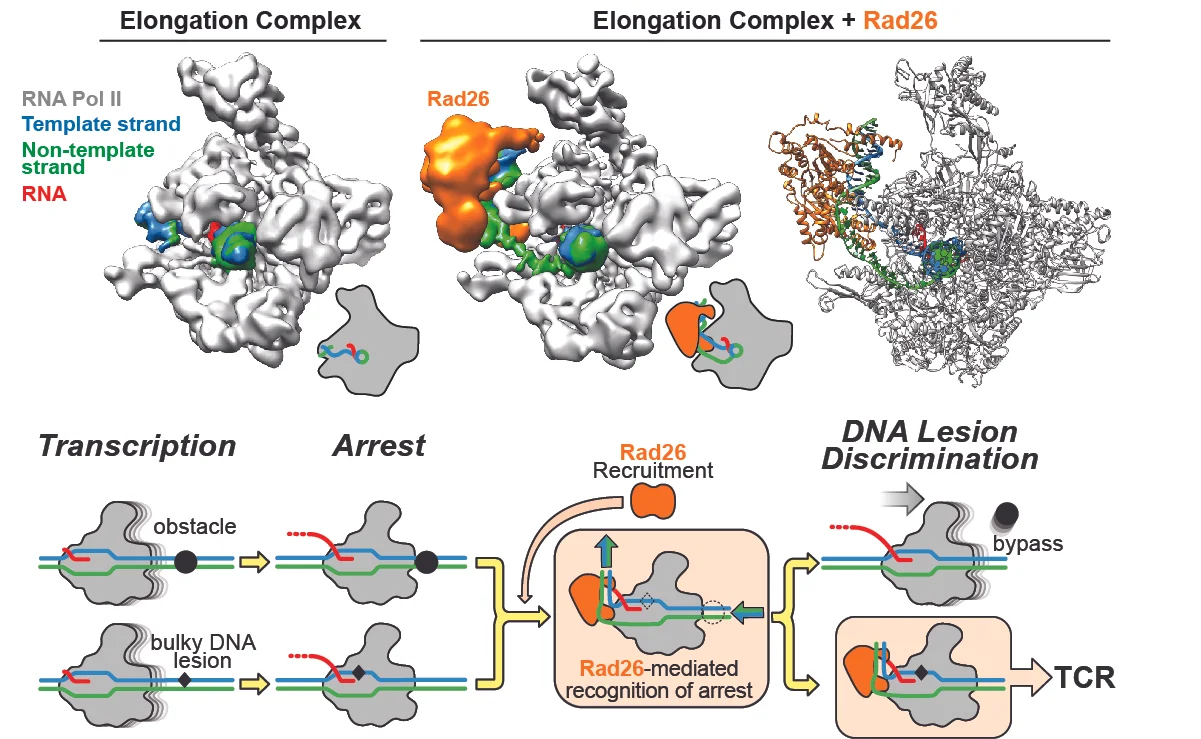
In our most recent work, in collaboration with Dong Wang's lab, we focused on Rad26, the yeast ortholog of CSB (Cockayne Syndrome protein B), a factor long known to be recruited to an RNA Polymerase II (RNPII) stalled at a site of DNA damage, where it initiates the process of Transcription-Coupled DNA Repair (TCR). Intriguingly, Rad26/CSB is a member of the Swi2/Snf2 family of nucleosome remodelers but if and how its remodeling activity was involved in TCR was not known. Using a combination of biochemistry and cryo-EM, we showed that Rad26 binds to the DNA behind RNPII (as well as to RNPII itself), and induces an almost 90o bend in the DNA. From that position, Rad26 uses its translocase activity to effectively pull the template strand away from RNPII, thus favoring forward motion by the polymerase (see Figure below). With this activity, Rad26 helps RNPII overcome obstacles that do not need to be repaired (such as stalling sequences or other blocks on the DNA), but it cannot help it bypass DNA damage, such as pyrimidine dimers. We proposed that this stalled complex, with its unusual structure, would recruit downstream TCR factors to repair the damage.
Publications
* Equal contribution
# Co-corresponding authors
Baker RW*, Reimer JM*, Carman PJ, Arakawa T, Dominguez R and Leschziner AE (2020). Structural insights into assembly and function of the RSC chromatin remodeling complex. bioRxiv.
Turegun B, Baker RW, Leschziner AE and Dominguez R (2018). Actin-related proteins regulate the RSC chromatin remodeler by weakening intramolecular interactions of the Sth1 ATPase. Comms. Biol. 1. (link)
Xu J*, Lahiri I*, Wang W, Wier A, Cianfrocco MA, Chong J, Hare AA, Dervan PB, DiMaio F, Leschziner AE# and Wang D# (2017). Structural basis for the initiation of eukaryotic transcription-coupled DNA repair. Nature 551: 653-7.
Nguyen VQ, Ranjan A, Stengel F, Wei D, Aebersold R, Wu C and Leschziner AE (2013). Molecular architecture of the ATP-dependent chromatin remodeling complex SWR1. Cell, 154:1220-1231. {download PDF}
Leschziner AE (2011). Electron Microscopy studies of nucleosome remodelers. Current Opinion on Structural Biology. 2011 21:709-718 {download PDF}
Leschziner AE, Saha A, Wittmeyer J, Zhang Y, Bustamante C, Cairns BR and Nogales E (2007). Conformational flexibility in the chromatin remodeler RSC observed by electron microscopy and the orthogonal tilt reconstruction method. Proc. Natl. Acad. Sci. USA 104(12): 4913-8. {download PDF}
Leschziner AE, Lemon B, Tjian R and Nogales E (2005). Structural studies of the human PBAF chromatin-remodeling complex. Structure (Camb) 13(2): 267-75. {download PDF}
CSB/Rad26 uses its Swi2/Snf2 translocase activity to help RNA Polymerase II overcome obstacles that do not need to be repaired, but cannot help it bypass DNA damage. The top row shows cryo-EM maps for an Elongation Complex alone (left) and bound to Rad26 (middle). The atomic model for the complex is shown to the right. Below is our proposed model for TCR initiation. Rad26 is recruited to stalled RNPII's, where its translocase activity can help it bypass certain obstacles. Because Rad26 cannot help RNPII bypass DNA damage, we propose that the unusual structure of the complex (with the Rad26-induced bend in the DNA behind the polymerase) would recruit TCR factors for DNA repair.